![]()
Genetic Linkage
Mendel's rule of independent assortment is not always true
- Bateson and Punnett first noticed this
- they crossed purple flower and long pea with red flower and round pea and did not get the expected 9:3:3:1 ratio
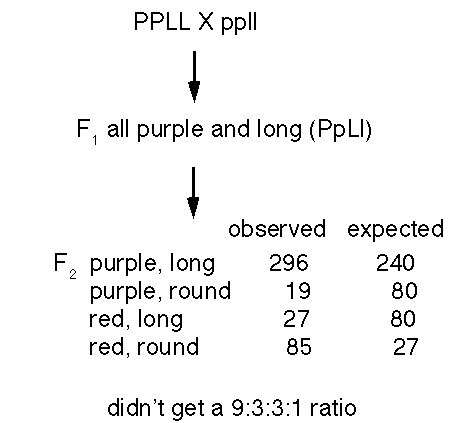
- T. H. Morgan - an excess number of progeny with the
parental genotypes
- he used a testcross of the F1 from either purple, vestigal X wild type or purple X vestigal

- dihybrid's gave different results depending on whether the recessive alleles were originally together - in which case they tended to stay together (coupling) or in separate strains - in which case they stayed apart (repulsion)
- Morgan's explanation: genes are on the same
chromosome and thus stay together unless
recombination occurs
- a linkage group is all of the genes that show linkage to one another
- the number of linkage groups is equal to the number of chromosomes
- If the genes were on the same chromosome, why were
all of the combinations showing up (albeit, sometimes at
very low levels)?
- Morgan proposed that the chiasmata observed during meiosis were the result of breaking and rejoining of the chromosomes, this produced recombinant chromosmes
- The frequency of recombinants might reflect the distance between the genes on the chromosome
Alfred Sturtevant pieced the maps of many different genes together to produce the first linkage map
Trihybrid mapping
Mapping with testcrosses of dihybrids is difficult
- It is hard to determine the order (need three different crosses)
- You can't measure the double recombinants so tend to underestimate distances
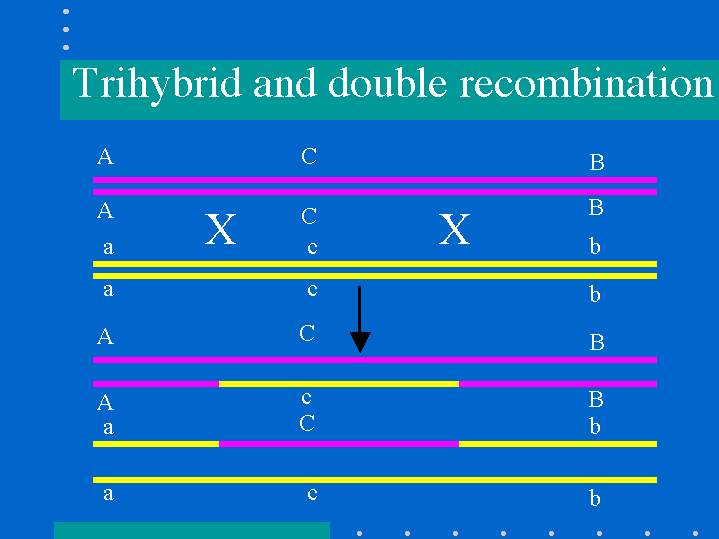
Solution: testcross of trihybrids (three-point mapping)
- can measure double recombinants
- allows direct determination of the gene order for the three genes
- can be used to calculate any interference between crossover events (coefficient of coincidence)
Missing Recombinants
- Eight different possible phenotypic combinations
- ABC, ABc, AbC. aBC, Abc, aBc, abC, abc
- The four reciprocal combinations(only one of each
phenotype) will have similar numbers
- ABC and abc, Abc and aBC, ABc and abC, AbC and aBc
- One reciprocal pair will be most frequent, this is the parental configuration
- One reciprocal pair will be least frequent, this is the pair that results from a double recombination (because two recombination events are less likely than one)
- The alleles that switch places between the parental and double recombinants are from the gene in the middle
Determining Distance
- Once the order is determined, adding the numbers of
one set of single recombinants to the double recombinants
and dividing by the total progeny will give the distance
from the middle gene to a gene on the end.
- If the parentals are ABC/abc and the doubles are ABc/abC then the order is ACB and (Abc + aBC +ABc + abC)/total is the distance from C to A
- Example Problem
Bell CSU Chico Library
This document is copyright of Jeff Bell
Last Update: Wednesday, August 12, 1998